1. 引言
在酶的六大类型中,30%~35%为氧化还原酶[1] ,由于氧化还原酶在催化制备手性醇、羟基酸、氨基酸方面显示出极大的优势,因此在制药、食品、精细化工、农药等领域具有重要的用途,它的应用也越来越受到人们的重视[2] 。但是大部分的氧化还原酶催化反应时都需要辅酶的存在,尤其是烟酰胺型辅酶[NAD(P)+和NADPH]的参与。然而辅酶大都价格昂贵,所以辅酶再生在工业应用上成为必须解决的问题。因此,根据生物催化反应的过程经济性和工业可行性,氧化还原酶在应用中除了必须有合适的酶和反应工程技术以外,还必须提供高效、低成本的辅酶再生系统。所谓辅酶再生,就是把辅酶从氧化态再生为还原态,或者反之,从而使辅酶保持在一定的催化剂量水平。此外,辅酶再生能够简化产物的分离,并有利于酶促反应向正反应方向移动[3] 。
近些年来,为了解决辅酶再生这一问题,已经提出了一系列的方法,主要有酶催化再生法和非酶催化再生法(包括化学再生法、电化学催化再生法和光化学催化再生法)[4] 。其中酶法再生的方法优势明显,反应速率快,选择性高,再生体系与合成体系兼容性好,过程易于监控。与其它用于辅酶再生的氧化还原酶相比,辅酶氧化酶具有以下优点:以O2为底物,可直接将NAD(P)H氧化为NAD(P)+,所以不需要添加新的底物;没有副产物产生,终产物水不会对与之耦联的酶产生抑制,不需额外引入其他底物和产生副产物。因此,辅酶氧化酶成为工业上用于NADH和NADPH再生的理想用酶。
本研究基于这样的背景,以生物信息学的研究方法,利用网络蛋白质数据库和各种软件,对辅酶氧化酶系进行分析对比,归纳总结辅酶氧化酶的分类,简要机理及其辅基周围重点氨基酸残基,重点分析烟酰胺型辅酶氧化酶的作用机理,蛋白质序列及其空间立体结构,通过蛋白质序列比对及3D结构比对获得活性中心所在位点,希望得到辅酶氧化酶的特征氨基酸序列,利于今后的蛋白质识别,得到活性中心及其辅基周围氨基酸残基,利于今后的酶的定向改造修饰等,也利用构建酶偶联系统进行辅酶的再生。
2. 材料与方法
2.1. 材料来源与工具
酶的序列及结构信息来源于网上共享的生物信息学数据库,如NCBI, PDB, SWISS-MODEL, SRS, ExPASy, Profile, InterProScan等。分析软件为ClustalW 2.0, ClustalX 2.0, PyMOL 1.0, VMD1.8.7和Rasmol 2.7。
2.2. 方法
应用Clustal序列比对软件对烟酰胺型辅酶氧化酶或脱氢酶的家族下的子集进行蛋白质序列分析,发现特征序列,辨别观察其同源性和保守位点。应用PyMOL 1.0和VMD1.8.7对获得的酶的空间结构进行分析,利用Swiss-Model在线系统分析酶的核心区域的立体结构,用软件Rasmol 2.7查看蛋白质的三维结构图。流程如图1:对NADH(P)H辅酶氧化酶的结构识别研究方法流程图。
3. 结果与分析
3.1. 序列比对
通过ExPASy的Enzymes数据库查得烟酰胺型辅酶氧化酶和脱氢酶都位于国际酶法分类中的1.6.-.-.这一子集中。除去该子集中没有蛋白序列及空间结构的酶,结果如表1所示。
应用ClustalX 2.0对Ec1.6.-.-下的每个子集的全体蛋白质序列进行全序列比对,得出每个子集中所有蛋白的序列比对图,观察其同源性和保守位点。图2是Ec1.6.99.3子集中的NOX的全序列比对的图(部分)。
本文选取Ec1.6.99.3子集为研究对象,Ec1.6.99.3子集下NADH脱氢酶一共有133种,其中有2种存在3D立体结构,存在于大肠杆菌和嗜热杆菌。该子集中包含我们重点研究的6种已命名辅酶氧化酶
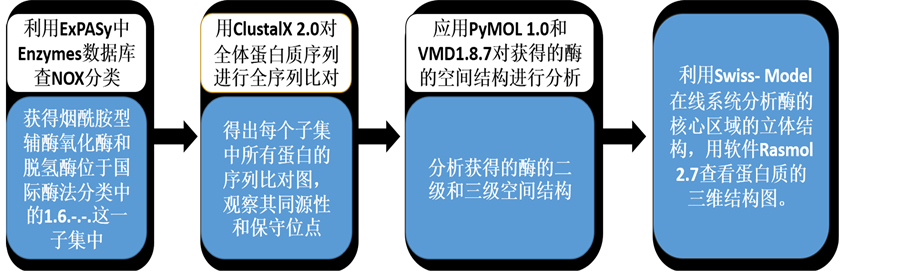
Figure 1. Flow chart of research method of the structure of coenzyme NADH(P)H oxidase identification
图1. 对NADH(P)H辅酶氧化酶的结构识别的研究方法流程图

Table 1. The number and name of enzyme under the subset of the international classification of enzymatic 1.6.-.- (last updated on May 19, 2010)
表1. 国际酶法分类下1.6.-.-子集下酶的编号和名称(2010年5月19日最后更新)

Figure 2. The whole sequence alignment of Ec1.6.99.3 (partial)
图2. Ec1.6.99.3的全序列比对(部分)
(NOX),其中P37061和A2RIB7为已知型NADH氧化酶,剩下的Q58065、Q49408、P75389,Q5XC60均为推断型NADH氧化酶。已命名的6种NOX的序列比对如图2,我们发现在10位和42位氨基酸左右残基高度相似。此外,我们同样发现每一子集中酶的同源性都很高,这说明酰胺型辅酶氧化酶或脱氢酶虽然在不同的生物中,但都催化同一类反应,有相同的电子接受体(图3)。
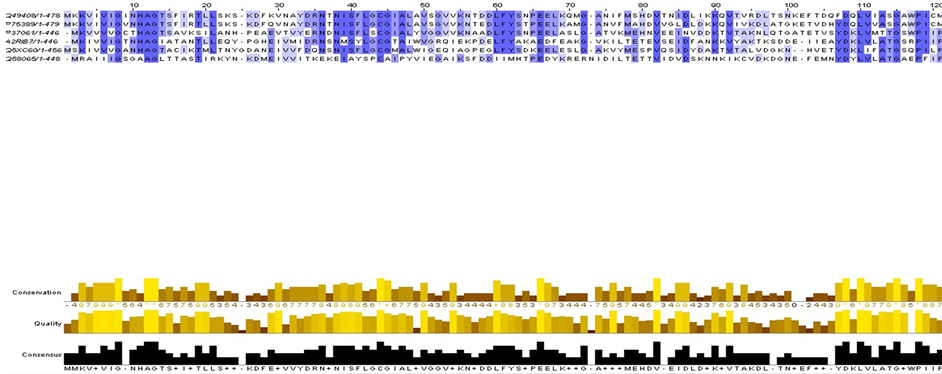
Figure 3. 6 named NOX protein sequence alignment in Ec1.6.99.3
图3. Ec1.6.99.3中6种已命名的NOX蛋白质序列比对
P37061存在于肠球菌中,全长446AA,可催化四个电子的氧气的还原反应。P37061的活性位点在Cys-42处,反应中氧化态Cys-SOH通过His-10的氢键帮助维持稳定。又根据PDB code:2cdu对其3D结构进行同源模建的,序列的同源度达55%,每个亚基都链接一个ADP和FAD辅基,FAD辅基以氢键和亚基结合,周围重点残基为Thr9, His10, Glu32, Csx42, Val81, Asp282, Ala300, ADP周围重点残基为Asp179, His181, Tyr188, Val214。
A2RIB7是存在于乳球菌中的NADH氧化酶,全长446AA,催化分子氧得到四个电子还原成水,可作用于beta-NADH,不作用于alpha-NADH,beta-NADPH或alpha-NADPH,在有氧的条件下,氧气是电子接受者,在厌氧的条件下,DCIPh和MB可以替代氧作为电子接受者。其3D结构是根据2bc0(PDBcode)进行同源建模得到的,序列同源度为49%。辅基为FAD,周围重点残基为Asn10, Asp34, Val81, Asp292, Ala310, Phe436。
3.2. 立体结构比对
通过一系列的蛋白质序列比对,我们观察到一个子集下酶的同源性较高,在序列比对过程中,可以看出一些氨基酸残基高度保守的位点,我们推测这些位点可能是酶的活性中心或辅基结合位点。由于每个子集的序列相似度较高,所以选取Ec1.6.99.3中存在已知蛋白质立体结构的2种已知型酶进行具体研究。先从酶的编号,功能开始,分析它的蛋白质二级结构,通过PDB下载其3D结构文件,利用PyMOL打开,再选取蛋白质亚基进行具体研究。根据之前的蛋白序列比对,选择包含辅基结合位点和活性中心位点的亚基。
图4、图5为P37061和A2RIB7这两种已知型NOX蛋白质二级结构域,比较发现在10位点为NOX的质子接受点,而42位点为其催化活性中心所在,并且在42位处紧密连接着一个FAD辅基。在明确这两个特殊位点后,又模拟了P37061和A2RIB7的立体结构(如图6),进一步考察这两个位点周围可能包含的蛋白质亚单位,结果显示活性中心周围的氨基酸残基呈现高度保守性,主要包含Gly43,Leu40或Gly43,Ser40这两类特征残基。为了验证这些位点的普遍性,我们又对两种推断型蛋白质Q58065、P75389进行同样的建模分析,结果,此种断型的NADH氧化酶活性中心也在42或43号氨基酸位置,该位置为Cys或Gly,周围出现Leu40或Ser40,所以通过3D结构比对可以推断出有这样保守结构的蛋白质序列极有可能是NAD(P)H氧化酶。
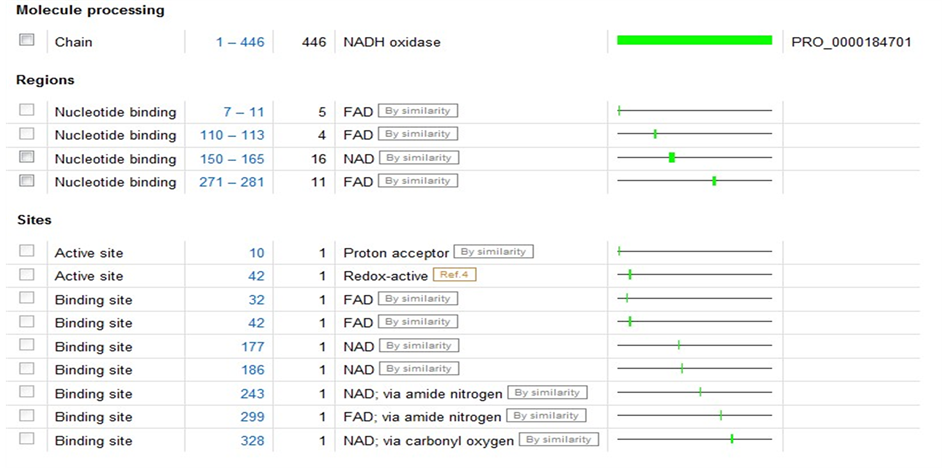
Figure 4. The secondary structure domain of protein P37061 (E. faecalis)
图4. P37061(E. faecalis)的蛋白质二级结构域
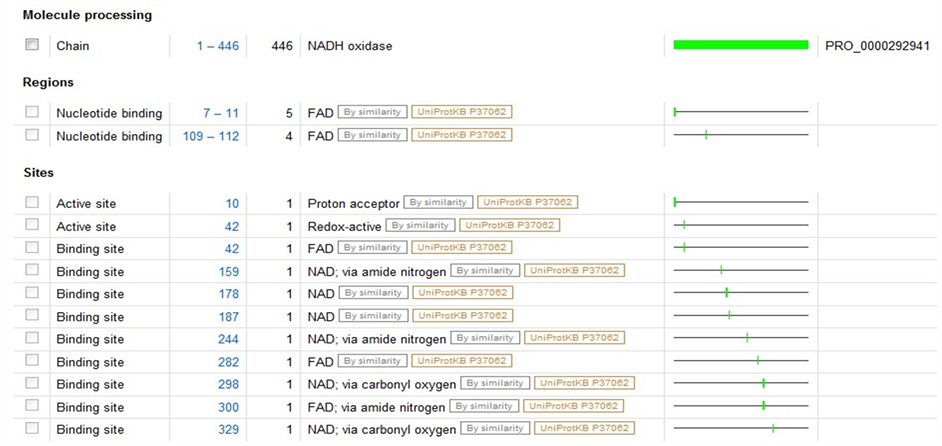
Figure 5. The secondary structure domain of protein A2RIB7 (L. lactis)
图5. A2RIB7(L. lactis)的蛋白质二级结构域
3.3. NOX结构功能域与催化机理
根据产物的不同,NOX可分为两类:一类产物为H2O2的,成为NADH过氧化酶或H2O2型NADH氧化酶(NOX-1),在生成H2O2的过程中发生了2个电子的转移[5] 。另一类产物为H2O的,就是通常所谓的NADH氧化酶(狭义)或H2O型NADH氧化酶(NOX-2),在生成H2O的过程中发生了4个电子的转移[6] 。
图7为辅酶氧化酶的典型结构域,主要分为NAD(P)H结合域,FAD结合域,ADP,FAD和二聚体域[7] 。FAD依赖型NADPH氧化酶可以等量的催化NADH或是NADPH氧化同时还原氧气生成水,稳态之后只有0.5%的还原产物被测定为是过氧化氢,这表明在酶氧化NADPH和氧气的反应之后,过氧化氢
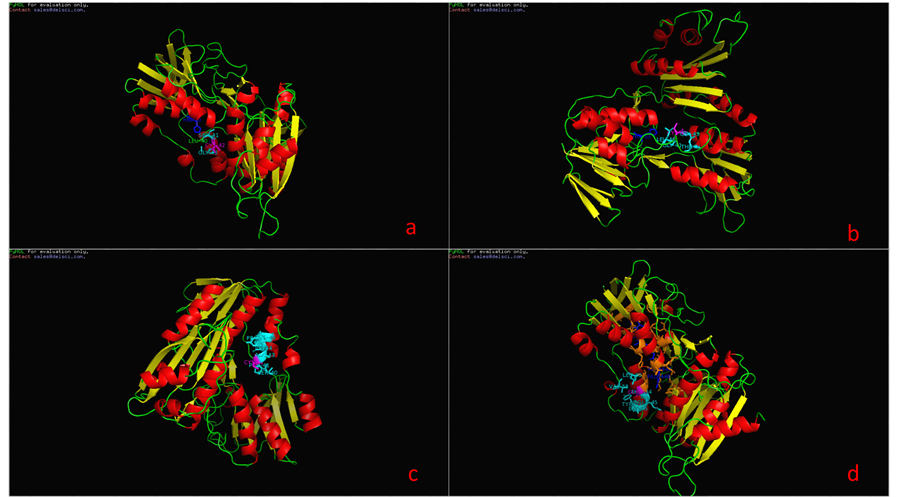
Figure 6. the catalytic center of loci and conservative amino acid residues of (a): P37061(E. faecalis), (b): A2RIB7 (L. lactis), (c): Q58065(M. jannaschii), (d): P75389(M. pneumoniae)
图6. (a):P37061(E. faecalis)、(b):A2RIB7(L. lactis)、(c):Q58065(M. jannaschii)、(d):P75389(M. pneumoniae)的催化中心位点及保守氨基酸残基
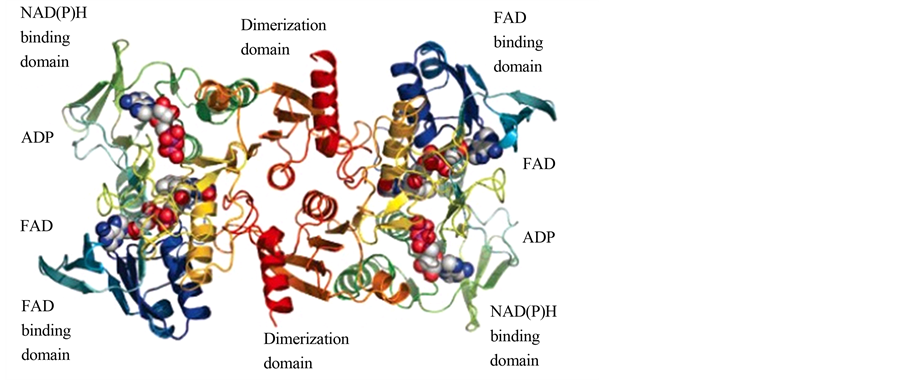
Figure 7. NOX protein molecule function domain (b1) [7]
图7. NOX蛋白质分子的功能域[7]
并没有离开这一系统。电子密度显示氧化还原活性中心Cys42邻近FAD的表面,可以被氧化为次磺酸形式(Cys-SOH)。Cys42的侧链也显示出两种构型,次磺酸以氢键与His10结合,另外一种构象是以氢键和FAD的氧原子结合。而且在NADPH结合区域还发现结合了辅基ADP,但它的存在并不影响酶的活性。根据电子密度图我们可以推测每一个NADH或NADPH底物都通过一条长长的通道从酶的外部一直延伸到FAD的内部[7] 。
通过上述对NOX催化活性中心位点的识别和对空间立体结构的分析,结合有前人对L. sanfranciscensis中NOX的研究[7] 可以总结出NOX催化中心的反应机理,NAD(P)H的氧化反应分为两部分,其中涉及了FAD的氧化还原,氧气被还原为过氧化氢,进而通过半胱氨酸的变化还原成水,还有NAD(P)H最终氧化成NAD(P)+,实现了烟酰胺型辅酶的再生。根据立体结构的显示,催化活性中心处的Cys42紧密连接FAD辅基,His10在其附近起电子接受体的作用,也帮助Cys42稳定其磺酸的氧化形式。机理显示,NAD(P)H反应时先和FAD结合,将其还原释放出一分子的NAD(P)+,氧气存在的情况下将氧气还原成过氧化氢,Cys42在过氧化氢还原成水的过程中起了重要作用,在这过程中它变成次磺酸形式,之后又通过FAD还原为原始形式,在这次FAD的构型变化中NAD(P)H又被氧化成NAD(P)+。
4. 结论
通过利用生物信息学的研究,对辅酶氧化酶系进行了总结和分类,了解其基本机理,先由Ec1.6.99.3子集下的P37061和A2RIB7这两种已知型NOX蛋白质序列比对,二级结构的考察,比较发现在10位点为NOX的质子接受点,而42位点为其催化活性中心所在。又对其立体结构的对比和分析,获得其催化中心的结构,分析发现活性中心周围的氨基酸残基呈现高度保守性,主要包含Gly43,Leu40或Gly43,Ser40这两类特征残基。根据又对两种推断型蛋白质Q58065、P75389进行同样的建模分析我们确定此性质具有一定的普遍性。所以通过3D结构比对可以预测推断有这样保守结构的蛋白质序列极有可能是NAD(P)H氧化酶。这可用于今后指导辅酶氧化酶的筛选,在分子层面修饰和改造酶的结构。今后的研究也可针对某一种辅酶氧化酶进行同源模建和分子对接,更能深入了解其作用机理和氨基酸残基对催化活性的影响。

NOTES
*通讯作者。