1. 引言
真核细胞中的DNA与有关蛋白质之间的连接称作染色质。染色质是所有遗传物质的载体,在生物群体中,染色质的结构特征具有相对的保守性,对不同种类生物的染色质进行分析研究能够揭示各种物种染色质的结构差异,从中反应出生物遗传的多样性及系统的演化路线。染色质有着紧密的组织结构,以至于染色质中大部分DNA序列不仅在空间构造上彼此不相接触,而且在功能上还处于沉默状态。
染色质结构的调节主要是通过三种方式实现的:DNA的甲基化,组蛋白的修饰,ATP依赖的染色质重塑复合物[1] 。染色质重塑复合物主要分为几个复合家族,它们通过特异地移动核小体组蛋白位置或者暴露核小体DNA区域以改变转录调节因子对DNA的可接近性,从而改变基因的表达调控 [2] - [4] 。
在所有真核生物中,染色质的基本亚基都是相同的,这些亚基又被称为核小体,无论是在分裂间期细胞核中的常染色质或异染色质中,还是在有丝分裂期的染色体中,核小体都是其中不变的组成成分,它提供了染色质的第一级结构组织方式。在核小体中,DNA与组蛋白的结合是动态的,并允许其它DNA结合蛋白间歇地靠近核小体上的DNA [5] - [8] 。核小体在基因组DNA分子上的精确位置称为核小体定位。
真核生物不同于原核生物的一大特点是具有染色质结构,DNA的序列信息被包裹在组蛋白结构中,基因的转录激活需要染色质结构的改变,染色质中与转录相关的结构变化称为染色质重塑,主要是指转录因子进入核小体,建立起有活性的启动子结构的过程。在转录过程中随着转录泡的移动,需要进行连续的染色质重塑,在此过程中重塑因子复合物的作用非常重要。染色质调节因子会作用于双链断裂处,对DNA损伤反应很重要的 [9] 。
Cell杂志报道了染色质重构调节子在核小体中特异性和方向性的最新研究成果 [10] 。研究人员分析了染色质重塑调节因子ISW2的作用机制。研究发现,染色质重构调节子在基因的起始和结束位置与特定核小体位点相互作用,并在移动中的核小体内,基因的5'和3'末端产生朝向或背向无核小体基因组区域的定向性。研究者发现在酿酒酵母细胞全基因组范围内,SWI/SNF,RSC,ISW1a,ISW1b,ISW2等染色质重构调节子复合体与单个核小体相结合,并通过缺失分析证实,这些复合体通过辅助核小体定位行使其功能。
ISW2复合物在营养生长期通过转录因子Ume6被招募到早期几个减数分裂基因的启动子区去抑制这些基因表达,ISW2复合物对减数分裂基因启动子的转录抑制与核酸酶不能接近染色体结构有关 [11] - [14] 。核小体通过ISW2来进行移动需要ATP酶与滑块区域一致行动,而且ISW2还涉及核小体排列和基因活性调控的规范化 [15] [16] 。本研究主要研究ISW2缺失条件下染色质重塑对核小体位置的影响。为了更加深入了解基因的功能,把YPD情况下的核小体的数据与WT、ISW2一起进行对比分析。
2. 方法和材料
2.1. 数据来源
我们的数据主要分为四个部分:第一部分是来自Lestyn Whitehouse等人测出的在WT和ISW2两种情况下两组核小体占位率数据 [17] (Shivaswan S. et al., 2007)。第二部分是来自Noam Kaplan等人实验合成的酿酒酵母体外核小体定位数据(YPD) [18] (Kaplan N. et al., 2009)。第三部分则来自NCBI数据库中的酵母16条染色体DNA编码序列。第四部分为David等人文献中给出的4792个高置信度酵母基因组实验数据(David L. et al., 2006),其中包括了转录起始位点(TSS)座标。
2.2. 数据处理
2.2.1. 数据预处理
YPD这组数据给出了转录起始位置到转录结束位置核小体的占位率数据,我们把没有占位率数据的位置设置为0 [19] 。WT和ISW2两种情况下核小体占位率的数据为每隔4 bp测量一次的数据,我们把没有测得数据的位置设置为0。按照16条染色体的长度设置三组16个空矩阵,将WT、ISW2、YPD这三组数据分别放入三组16个空矩阵中,就可以分别得出三组16条染色体上核小体占位率数据。
2.2.2. 对齐处理
以David等人所提供(David L. et al., 2006)的高置信度转录实验数据中给出的4792条基因的TSS为参照坐标,我们分别将上述不同实验平台和实验条件下得到的三组酵母16条染色体核小体实验数据,以4792条基因的TSS为中心,选取上、下游各800 bp数据进行了对齐(考虑到基因的方向性,对于C型基因所对应的数据还要进行镜像反折),然后将这些数据进行叠加、平均、平滑及归一化处理,得到了以基因TSS对齐的核小体平均占位图谱,如图1所示。
由图可以得出三种情况下核小体位置的变化,如表1所示。
由表1可以看出以WT核小体位置为基准,ISW2缺失情况下核小体位置有变化,ISW2缺失相对于WT的位置是远离TSS的,所以在ISW2缺失的情况下,核小体的位置是远离TSS。YPD情况下相对于WT变化规律不明显,可以观察到在TSS下游YPD相对于WT的位置的远离TSS。而且YPD的占位率明显比另外两种情况要高。
2.2.3. K均值聚类
K均值聚类是先随机选取K个对象作为初始的聚类中心。然后计算每个对象与各个种子聚类中心之间的距离,把每个对象分配给距离它最近的聚类中心。聚类中心以及分配给它们的对象就代表一个聚类。一旦全部对象都被分配了,每个聚类的聚类中心会根据聚类中现有的对象被重新计算。这个过程将不断重复直到满足某个终止条件。
为了更加进一步了解三种情况下的基因的功能,用K均值聚类的方法对对齐后的三种占位率数据进行聚类,经过多次试验结果,聚为20类为最佳聚类,聚类后的20种核小体占位率情况可以分为5大类。第一类:−1核小体不存在;第二类:−1核小体不明显;第三类:+1核小体不明显;第四类:核小体位置不变;第五类:核小体位置不明显。图形如图2所示。
图2中核小体的位置变化以WT和ISW2缺失条件下的核小体位置为基准,核小体的位置移动是YPD相对于WT和ISW2缺失两种情况下核小体位置的变化。将4792个基因名称导入SGD数据库中得到了75种基因功能,为了进一步了解基因的功能,将上述五种情况的基因分别与75种功能的基因进行比对,把相同的基因提取出来,并且计算出相同基因的个数,得出的结果如表2所示。
分析后发现有2种基因功能可能会影响−2,−3,+2,+3核小体形成,这2种基因功能分别为:核糖体的结构成分(structural constituent of ribosome),磷脂酰肌醇磷酸结合(phosphatidylinositol phosphate binding)。有6种基因功能可能会影响−1核小体的形成,这6种基因功能分别为:mRNA结合(mRNA binding),泛素蛋白转移酶活性(ubiquitin-protein transferase activity),ATP酶活性,耦合(ATPase activity,
 注:其中黑色曲线表示WT条件下对齐后的占位率图谱,红色曲线表示IWS2缺失条件下对齐后的占位率图谱,绿色曲线表示YPD条件下对齐后的占位率图谱。
注:其中黑色曲线表示WT条件下对齐后的占位率图谱,红色曲线表示IWS2缺失条件下对齐后的占位率图谱,绿色曲线表示YPD条件下对齐后的占位率图谱。
Figure 1. The map of nucleosome occupancy under three case
图1. 三种情况下核小体占位率图谱
 注:图2中绿色线表示聚类后YPD条件下核小体占位率的图谱,黑色线表示聚类后WT条件下核小体占位率的图谱,红色线表示聚类后ISW2缺失条件下核小体占位率的图谱。
注:图2中绿色线表示聚类后YPD条件下核小体占位率的图谱,黑色线表示聚类后WT条件下核小体占位率的图谱,红色线表示聚类后ISW2缺失条件下核小体占位率的图谱。
Figure 2. The map of nucleosome occupancy under five case
图2. 五种情况下核小体的占位率图谱

Table 1. Three kinds of nucleosome center position
表1. 三种情况下核小体中心位置

Table 2. The same number of genes which compared with five types of genes and seventy-five kinds of gene function
表2. 把五种类型基因与75种基因功能比较有相同基因的个数
coupled),S-腺苷甲硫氨酸依赖的甲基转移酶的活性(S-adenosylmethionine-dependent methyltransferase activity),基底的特异性转运蛋白活性(substrate-specific transporter activity),转运活性(transporter activity)。这6种基因功能可能会抑制−1核小体的形成。有3种基因可能会影响+1核小体的形成,这3种基因分别为:转录因子结合(transcription factor binding),转移酶活性,转移含磷基团(transferase activity, transferring phosphorus-containing groups),转移酶活性,转移酰基(transferase activity, transferring acyl groups)。其中转录因子结合(transcription factor binding)可能抑制+1核小体的形成,转移酶活性,转移含磷基团(transferase activity, transferring phosphorus-containing groups)和转移酶活性,转移酰基(transferase activity, transferring acyl groups)可能促进+1核小体的形成。
2.2.4. 碱基Logo
我们为了更加深入了解基因的功能,对以上11种基因功能中的9种基因功能的基因分别画出碱基图,对−1核小体有影响的就以WT−1核小体的位置为基准前后各取15 bp共31 bp的碱基画图谱,同理对+1核小体有影响的以WT+1核小体位置为基准前后各取15 bp共31 bp的碱基画图谱。以下图谱仅分析对−1核小体和+1核小体的影响,图谱结果如图3~11所示。
由图3~8可以看出−1核小体周边的碱基以TTT或AAA为多,TTT与AAA的硬度为−0.274属于硬度较强的密码子 [20] ,一定程度上可能会抑制−1核小体的形成,有的碱基不明显但硬度偏硬的密码子比较多,从而抑制了−1核小体的形成。
Figure 3. mRNA binding base Logo map
图3. mRNA binding碱基Logo图谱
Figure 4. Ubiquitin-protein transferase activity base Logo map
图4. Ubiquitin-protein transferase activity碱基Logo图谱

Figure 5. ATPase activity, coupled base Logo map
图5. ATPase activity, coupled碱基Logo图谱

Figure 6. S-adenosylmethionine-dependent methyltransferase activity base Logo map
图6. S-adenosylmethionine-dependent methyltransferase activity碱基Logo图谱
Figure 7. Substrate-specific transporter activity base Logo map
图7. Substrate-specific transporter activity碱基Logo图谱

Figure 8. Transporter activity base Logo map
图8. Transporter activity碱基Logo图谱
Figure 9. Transcription factor binding base Logo map
图9. Transcription factor binding碱基Logo图谱
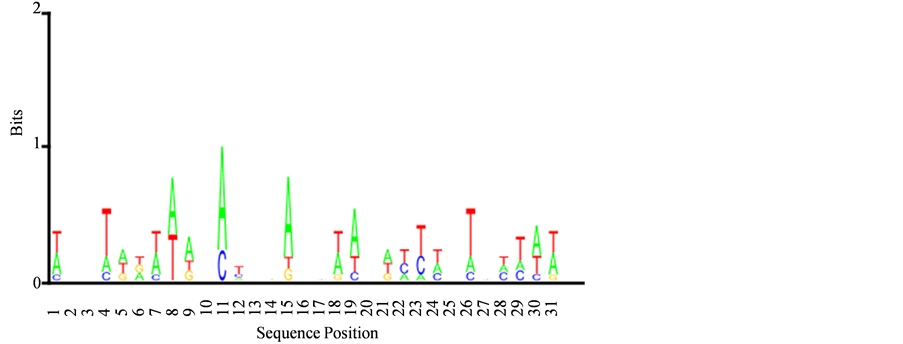
Figure 10. Transferase activity, transferring phosphorus-containing groups base Logo map
图10. Transferase activity, transferring phosphorus-containing groups碱基Logo图谱

Figure 11. Transferase activity, transferring acyl groups base Logo map
图11. Transferase activity, transferring acyl groups碱基Logo图谱
由图9可以看出碱基以A偏多,其中可以明显看到AAA和AAT这两种密码子,AAT的硬度为−0.28,属于硬度最强的密码子,从而抑制了+1核小体的形成。
由图10和图11可以看出,有多种密码子,而且密码子的软硬度不同,以硬度较软的密码子比较多,从而促进+1核小体的形成。
3. 结论
对核小体的位置移动的研究发现,在ISW2缺失的情况下,核小体的位置相对于WT是远离TSS的。在YPD情况下位于TSS下游核小体的位置相对于WT的远离TSS。对基因功能的研究发现有11种基因功能可能会影响核小体的形成。研究发现这11种基因功能中有9种基因功能可能是作用于−1核小体和+1核小体,该结果与Iestyn Whitehouse等人研究的结果ISW2作用于启动子邻近区域相符,说明ISW2缺失情况下可能会影响9种基因功能对核小体的定位。
基金项目
国家自然科学基金资助项目(31160234);云南省应用基础研究计划项目(2011FB082);省高校重点实验室开放基金(MWK201104);云南民族大学校级研究生一般项目(2015YJCXY255)。