1. 引言
肥胖,即脂肪组织的过量沉积,是目前工业化国家面临的首要健康问题。成脂过程中,间充质干细胞(MSC)分化成为脂肪细胞,最终导致脂肪细胞数量增多,体积增大和脂质在脂肪细胞中的积累 [1] 。大量的实验研究已经揭示了成脂过程中的一些重要的分子机制。许多研究显示PPARg是脂肪生成的关键调节因子 [2] - [4] ,其次,一些碱性亮氨酸拉链分子,如C/EBPa、C/EBPb和C/EBPd也参与诱导脂肪生成,并通过PPARg-C/EBPa正反馈循环在终末分化阶段扮演重要角色 [5] [6] 。另外,越来越多的研究者也开始关注microRNA在成脂调控中的作用,研究显示miR-21 [7] 、miR-27a [8] 、miR-143 [9] [10] 和miR-211 [11] 等均参与成脂关键基因的转录后调控。
在生物医学领域,通过文献整合信息的过程通常分为3步:1) 在公共数据库(如:PubMed、WOS)通过关键词检索出一系列相关文献;2) 人工查看这一系列文献获取一系列相关基因或者其他信息;3) 如果这些文献的信息不够充分,再进一步到各种专业数据库(如:GO)检索其他相关信息。在这种情况下,一个研究者要想通过传统的方式提取到需要的信息,就必然面临如下挑战:在阅读大量文献和通过数据库查询信息的过程中,花费大量时间,同时错失掉大量有用信息;其次,不同研究者提取信息的过程中,也出现了大量的重复性劳动。而文本挖掘(Text-mining),为简化我们的阅读过程,自动提取文献信息提供了一种可能 [12] 。
现有的数据库已关注到转录因子 [13] [14] 、microRNA [15] 、蛋白–蛋白互作 [16] 和表观遗传因子 [17] 等调控因子各自的调控关系网,然而,由于缺乏一个对数据进行综合加工处理的信息中心,各数据库信息之间的交互情况尚不清楚。其次,虽然文献的大量发表催生了许多新的信息检索技术的发展,但是,这些检索系统通常是以文档为单位进行检索,而非研究成果的内容。
在本研究中,我们搭建了一个公开的,可通过网页访问的,提供成脂分化研究最新动态的信息中心——ARN数据库平台。我们不仅收集和挖掘了与成脂相关的文献信息,还从专业角度对数据进行重新组合,消除冗余并通过外部数据库填补缺失的信息。除记录了实验已验证的数据和调控关系,还收集了与成脂相关的大量候选基因。此外,我们还通过可视化技术对调控网络进行展示,将各种参数,如节点的分类、功能、试验方法和材料等在网络中进行可视化标记。
2. 材料和方法
2.1. 数据来源
ARN数据库所有数据和文献均来自NCBI-PubMed、NCBI-Gene、miRBase、miRGate、PAZAR和TRRUST六个数据库,通过文本挖掘技术辅以专业人员的人工检查、注释和数据交互整合所得。
2.2. 数据库的构建
该数据库从数据采集到加工处理的流程主要包括以下四步(图1)。
第一步,文献信息的采集与挖掘。1) 文献采集。借助Cytoscape数据分析平台的Agilent Literature Search文献检索插件 [18] ,分别将47个已知的与成脂分化调控相关的基因 [19] (包括PPARg, CREB, C/EBPa, C/EBPb, C/EBPd, TGF-beta, GR, GATA2, GATA3, RHO, ROCK2, STAT3, KLF4, GEFT, YAP1, KROX20, TZA, BMP2, REV-ERB-alpha, EPAC,SMAD1, HDAC9, SHN2, BMP4, EZH2, SMAD3, TCF7L1, SIRT1, SIRT2, ZFP423, SETD8, MMP14, WNT10b, SETDB1, WNT5a, STAT5a, PTIP, JAK2, RAC, TIMP3, ASXL1, MLL3, JMJD2C, PKA, ASXL2, WNT5b, Nocturnin, TLE3)与“adipo* differen*”提交到PubMed,
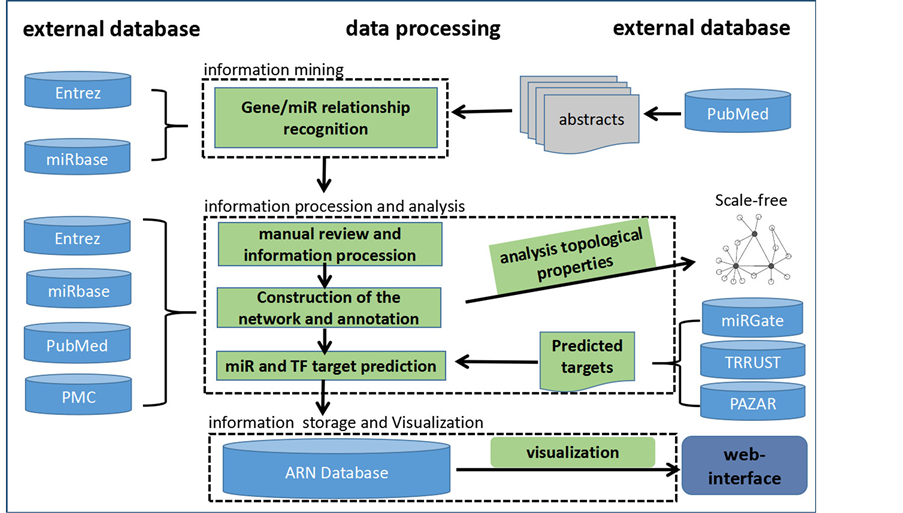
Figure 1. Database construction pipeline
图1. 数据库构建流程图
进行联合检索,在进行47轮检索后,总计得到9908篇PubMed数据库的文章摘要作为初始语料库用于进一步分析。2) 文献挖掘。Agilent Literature Search首先将检索得到的文献拆分成单个的句子,再根据基因数据库(Entrez Gene)的基因名称或别名(概念名词),或者特定的关系词(通常是动词)如“binding”或“activate”进行分析,如果在一个句子中同时存在至少两个概念名词和一个关系动词,那么它就会被转换成一条互作记录,并加入到Cytoscape数据分析平台的互作网络当中。
第二步,人工筛选,注释和分析。1) 人工筛选。文献挖掘方法共同存在一个基础性的问题,多义词:即同一个词可能同时含有多个语义或归属于不同的分类 [20] 。这类问题在生物医学文献中很常见。例如:fat,可以是名词“脂肪”,也可以是形容词“肥胖的”,二者在生物医学类文献中均常常使用。在PubMed中检索“fat”,返回187,888个结果,你会发现fat同时还用于命名基因,或者作为通用符号使用。因此,有必要对文献挖掘的结果进行人工的检查,删除错误的联系。2) 人工注释。为了完整呈现调控网络的相关信息,专业人员根据文献摘要对特定基因,microRNAs和它们之间的关系进行了注释,标记出节点的分类、功能以及它对脂肪生成的影响,我们还对每篇文献的试验设计的相关信息如试验方法、试验材料、细胞系名称等进行分类整理。3) 设计分析指标,度量节点的重要性。在获得了大量的成脂分化相关数据后,我们发现基因或microRNA对成脂分化的重要性体现在3个方面:a) 它在成脂分化相关过程中表达量变化较大;b) 它与成脂分化相关的节点关系紧密(调控或者被调控);c) 它可能与成脂分化相关的节点关系紧密(有预测关系,待试验验证)。因此我们将IFi作为评价节点重要性的综合指标,结合互作关系数量、表达记录数量和靶标预测数量,利用IFi = 1/3(Ri/Rmax + Ei/Emax + Pi/Pmax)*100%公式计算出每个节点的IF [21] 。
转录因子和microRNA是成脂分化调控中重要的两类调控因子,因此我们通过TRRUST (http://www.grnpedia.org/trrust/) [13] 和PAZAR (http://www.pazar.info/) [14] 收集了所有可能与成脂相关的转录因子–靶标关系。通过miRGate (http://mirgate.bioinfo.cnio.es/miRGate/) [15] 收集了microRNA-靶标关系,为了确保记录的可信度,我们只保留了预测数大于3的结果。
第三步,信息的储存与可视化。ARN数据库是运用Microsoft SQL Server关系型数据库平台开发的,Web界面是运用.NET和HTML5开发的,用户可以通过浏览器访问ARN数据库平台(http://210.27.80.93/am/);成脂调控网络的可视化是通过D3(d3js.org)实现的。
第四步,动态分析工具的设计。动态网络分析(DNA)是一个新兴的科学领域,汇集了网络技术和理论中的社交网络分析(SNA),链接分析(LA)等技术和概念。在成脂调控网络中有多种类型的节点(多节点)和多种类型的关系,这样的网络一般被称为高维网络。鉴于成脂调控网络是动态的,多节点的,多维的网络,ARN数据库的分析工具为用户提供了筛选功能,用户可根据节点参数设置过滤条件,从多个维度同时筛选。
3. 结果
3.1. ARN数据库网站页面组成
ARN数据库网站为用户提供地图页面、节点页面、文献页面、表达页面、分析页面和下载页面,用户可根据信息种类自由选择。该数据库的主页显示了成脂的可视化调控网络,其中包括了连接数最多的300个节点,用户可以通过“Node Number”选择页面显示的节点数(≤300)。节点的颜色和形状由其分类和功能决定,连线的颜色和形状由节点间的互作方式决定(图2)。
节点页面包含六部分内容(图3)。① 是基因或microRNA的一般信息;② 是基因或microRNA的相关文献概述;③ 是节点在不同条件下的表达情况;④ 是节点的单核苷酸多态性;⑤ 是一个可视化网络关系图,包含其对成脂的潜在影响、互作方式及试验材料或方法等信息;⑥ 是转录因子和microRNA

Figure 2. Screenshot of the home page
图2. ARN数据库首页示例
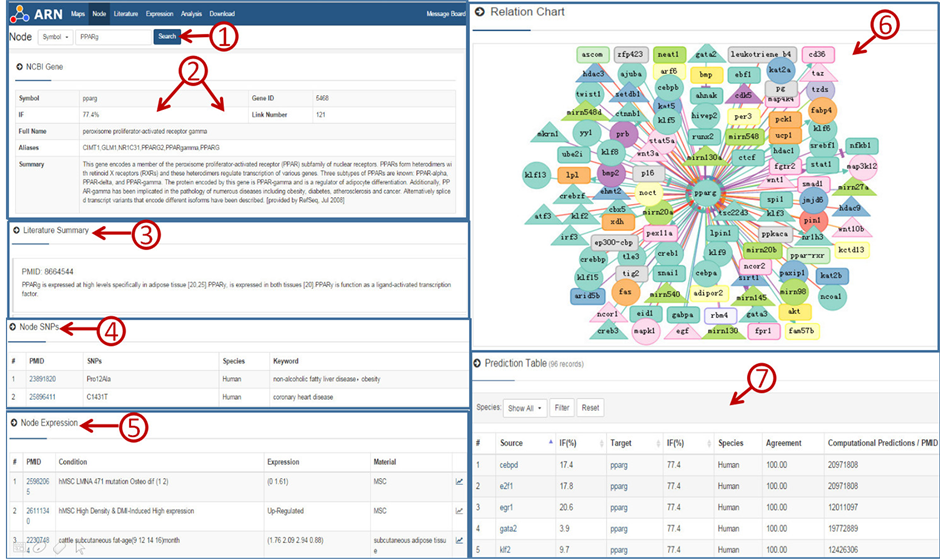
Figure 3. Screenshot of the PPARg results page
图3. PPARg检索结果页面示例A
可能的预测靶标关系,新的预测关系用黑色加粗线条表示。
地图页面(图4(B))展示了一些综述成脂分化的文献中的经典关系图。分为转录调控因子、信号通路因子、表观遗传修饰因子、非编码小RNA、环境因子和细胞生长因子六类,每张关系图下面以表格形式列出了关系图中所有节点,用户可通过点击节点基因符号,链接到特定基因的节点页面,我们还为用户提供了这些节点的互作网络图。
文献页面(图4(C))提供了综述、研究性文献,单核苷酸多态性相关文献和高通量分析文献四类文献的基本信息,并进一步整理了每篇文献所用的材料和方法。
表达页面提供了成脂分化过程中相关基因的表达情况(图4(D))。通过检索基因,用户可以获得同一个节点的收集自不同文献的表达数据,并查看表达变化的折线图。
分析页面是基于ARN数据库平台中的所有数据,让用户针对特定功能基因和microRNAs的数据进行分析和筛选。
我们还为用户提供一个下载页面(图4(E)),用户通过设置相关参数,可筛选出满足特定条件的基因ID,官方名称和相关文献的PMID,这些信息在其他数据库可直接使用。
同时,ARN数据库还包含一个留言板页面,让用户能随时指出数据库的错漏,以利于我们及时改进。
3.2. ARN数据库方便用户根据研究需要进行灵活检索
ARN数据库为用户提供多种方式进行灵活检索。基础信息的检索可以通过基因的ID号或官方名称、MicroRNAs的成熟序列号和PubMed数据库文章的PMID号进行;针对数据的特点,我们还为表达页面和文献页面设计了高级检索功能。
3.3. ARN数据库方便用户根据研究需要进行定制分析
目前,该数据库从1457篇与成脂相关的文献中共收集到与成脂相关的3054个节点,1807条互作关系,10,675条表达记录,1141条概述记录,43张成脂调控网络图。此外,我们还通过miRGate、PAZAR和TRRUST为ARN数据库的所有节点确定了12,696条可能与成脂相关的调控关系。基于以上所有数据,用户可以在分析页面,设置参数和筛选条件对特定节点、高通量文章或节点集进行定制分析。
4. 讨论
4.1. 少而精的专业信息中心
当我们在NCBI PubMed数据库中检索adipo*differe*时,会得到3万多条结果,NCBI PubMed 数据库会为每篇文献添加上物种、期刊名称,出版日期和关键字等信息,用户只能根据这些内容检索并分析。而在ARN数据库中,文献总量虽不到PubMed的1/10,但我们为每篇文章添加了更详细的信息,如所用细胞系的名称、样品种类等,方便用户进行更多样的检索和分析。图5中,我们提供了物种、细胞系和试验样品材料的统计数据、常用试验方法以及ARN数据库中的不同类节点的关系数量。在实际应用中,用户可能只关心其中的一部分信息,细节信息的添加使用户可快速筛选出高契合度的目标文献。
4.2. 节点信息的多样性
ARN数据库为每个节点添加了“分类”、“功能”、“IF”、“表达”和“分化方向”等内容,方便用户快速查找自己感兴趣的信息。在实际操作中,当我们在NCBI PubMed中检索“PPARg”时,会出现900多条单一记录,但当我们在ARN数据库检索时,虽仅有218条记录,但是汇集了NCBI-Gene,miRBase,NCBI-PubMed,miRGate,PAZAR,TRRUST等六个网站的151篇论文信息。从PPARg基因

Figure 4. Screenshot of the PPARg results pages B-E
图4. PPARg检索结果页面示例B-E

Figure 5. Basic statistical data of ARN database
图5. ARN数据库的基础统计数据
的基本信息,表达变化水平(Node Expression),调控关系网络(Relation Chart)到调控关系预测图(Prediction Chart),用户可多元化的了解该基因的详细信息,另外预测图还可帮助用户识别PPARg的边际信息,通过节点“IF”的排序,筛选出最重要的靶标预测关系,表1给出了部分预测结果,深入分析该表,我们得知Pan YC等 [22] 在进行癌症细胞的凋亡研究时,得出e2f1和cebpd都对PPARg进行转录调控,由于这三个节点都参与调控成脂分化过程,因而研究人员可以根据该结果,进一步设计试验研究该调控关系对于成脂分化的影响。
4.3. 数据的重组与扩展
大量数据的整合往往会产生新的信息,随着大数据技术的发展,数据的整体会比局部更具价值。如果我们综合分析多个数据集,很可能会发掘出有潜在价值的隐藏信息,最大限度的释放数据的潜在信息能量。例如,图6中,当我们把TRRUST数据库 [13] 中的所有8000多条记录所包含的748个转录因子和2374个靶基因分别与ARN数据库中的2000多条节点记录,进行交互分析时,得到了3538条成脂相关的转录因子靶向调控关系,它们所包含的转录因子和靶标均在ARN数据库中有记录。将来,当出现其它专业的调控因子数据库时,通过将它与ARN数据库进行交互,我们能够短时间内将其中可能与成脂分化相关的数据批量添加到ARN数据库平台。

Table 1. Top 10 PPARg prediction results
表1. PPARg部分预测结果
本表只给出部分预测结果,完整的数据集见ARN数据库。

Figure 6. Venn diagram to represent the overlap between ARN database and TRRUST database
图6. ARN数据库与TRRUST数据库记录的交互分析韦恩图
4.4. ARN数据库的改进和完善
脂肪细胞的前体细胞间充质干细胞(MSC),也能分化为成骨细胞,软骨细胞或成肌细胞 [23] ,所以涉及MSC定向分化的调控因子的研究将会对人类肥胖、骨质疏松和再生医学等健康问题的解决奠定科学基础。因此,我们将添加更多有关间充质干细胞的定向分化的相关信息和数据。其次,近两年来有关长链非编码RNA (lncRNA)参与调控成脂分化的研究 [24] [25] ,也获得了大量新的数据,一旦这些信息得以公开,我们会尽快将其补充到ARN数据库。另外,根据ARN数据库相关文献的作者机构信息,我们将很快实现机构合作信息的可视化,这将有利于科研思想的跨平台交流,机构间的项目合作和资源共享。
5. 结论
ARN数据库的数据和文献信息整合分析自多个公开数据库,该数据库是一个有关成脂分化的简单快捷的在线检索分析工具。我们计划每周更新数据库,以提供最新资源,紧跟成脂领域的研究前沿。期望该数据库能成为研究成脂分化的研究人员交流信息和孵化科研猜想的平台,促进成脂分化的机制研究。同时也希望ARN数据库能够启发研究者开发出更多的专业性数据库,让公共数据库信息的潜在价值得到最大释放。
基金项目
国家自然科学基金(批准号:31272411)、国家“863”计划(2013AA102505, 2011AA100307-02)资助。