摘要:
为了对比线粒体16S rRNA和COI基因片段在长江口虾虎鱼科鱼类种类鉴定和系统分类研究中的适用性,本文运用PCR技术,扩增了虾虎鱼科7属8种24个个体的线粒体16S rRNA和COI基因片段,并对其种间的序列差异进行比较分析。经比对后得到16S rRNA基因长度为535bp的序列,共编码178个氨基酸,24条16S rRNA序列共有12个单倍型,共检测到变异位点161个,约占总位点数的30.1%,插入/缺失位点15个,转换与颠换比值(Si/Sv)为1.51;同时,获得COI基因长度641 bp,共编码213个氨基酸,单倍型个数与16S rRNA序列相同,变异位点221个,所占百分比高于16S rRNA序列(约34.5%),无插入/缺失位点,转换/颠换值为1.29。两种基因的转换/颠换值均小于2,因此对其做了相应的突变饱和性分析,结果显示,两种基因在虾虎鱼科鱼类中不存在突变饱和现象。遗传距离结果显示,16S rRNA基因种间序列变异程度均小于COI基因,16S rRNA基因的种内和种间平均遗传距离分别为0.002和0.169,种间约为种内遗传距离的84倍,种间遗传距离最小值存在于纹缟虾虎鱼和髭缟虾虎鱼之间为0.100;而COI序列的种内和种间平均遗传距离分别0.001和0.215,种间为种内遗传距离的215倍。因此,16S rRNA和COI序列均适用于虾虎鱼科的种类鉴定。基于16S rRNA和COI基因序列获得的UPGMA系统树显示虾虎鱼科鱼类均可形成单系群,不同之处在于16S rRNA基因序列主要表现种、属间序列差异,而COI则突出表现种、属、科间序列差异。因此,在应用这两种基因片段做系统分类研究时,应根据研究的不同系统水平选择适当的分子标记。
Abstract:
In order to compare the applicability of mitochondrial 16S rRNA and COI (mitochondrial cytoch-rome oxidase subunit I) in species identification and phylogeny of Gobiidae from Yangtze Estuary, this study investigated the genetic characteristics of these two gene fragments. The two gene fragments of 8 species in Gobiidae were amplified by PCR, and the sequence variation among in-terspecies was analyzed. A total of 535 bp, 12 haplotypes, 161 variation sites (about 30.1% of all), 15 insertion or deletion of sites were detected for 16S rRNA of 24 individuals, and 178 amino acids were encoded. All of the transitional pairs (si) was slightly more than that of the transversional pairs (sv), and the ratio (R = si/sv) was 1.51; while 641 bp and 221 variation sites were detected for COI, and no insertion or deletion of sites was found in the COI gene, the ratio of si and sv was 1.29. Both the ratios of the two genes from 24 individuals were less than 2, so mutation saturation of the two genes was analyzed. However, the result showed that there was no mutation saturation in the two genes of Gobiidae. The results of genetic distance showed that the degree of variation of 16S rRNA in interspecific sequence was less than that of COI sequence. Intraspecific and interspecific K2P genetic distances were respectively 0.002 and 0.169 for 16S rRNA sequences, and 0.001 and 0.215 for COI sequences. Genetic differences of 16S rRNA sequences between congeners were, on average, about 84 times higher than the differences within species, and the minimum value (0.100) was existed between Tridentiger trigonocephalus and Tridentiger barbatus; while genetic differences of COI sequences were 215 times between intraspecies and interspecies, which met Hebert’s species identification standard (more than 10 times the difference between intraspecies and interspecies). Therefore, 16S rRNA and COI sequences were both applicable for species identi-fication in Gobiidae. The UPGMA phylogenetic tree was constructed based on 16S rRNA and COI, which demonstrated that Gobiidae in the Yangtze Estuary was a monophyletic group. The major differences were that interspecies and genera were more emphasis for 16S rRNA gene, while it highlighted interspecies, genera and family more for COI gene. As a result, it is advisable to select appropriate molecular markers according to different classified levels when applying the two gene fragments for studying species identification and phylogeny.
1. 引言
虾虎鱼科(Gobiidae)鱼类隶属鲈形目、虾虎鱼亚目,是现存的鲈形目鱼类中最大的一个类群,中国现有虾虎鱼类9科5亚科106属307种,约占世界虾虎鱼类总数的14% [1] 。虾虎鱼类是以暖水性为主的海洋或淡水小型鱼类,主要以摄食小鱼和虾为主,是中华鲟幼鱼索饵洄游的主要饵料生物,在长江口生态系统中具有重要地位 [2] 。虾虎鱼类体态变异大,体型微小,其早期形态存在较多相似性,在传统形态学鉴定中存在较多困难,导致其在种类的命名及系统分类关系上存在较大争议。目前,线粒体基因由于其具有结构简单,进化较快等优点,在动物种群的种质鉴定和系统分类研究中具有重要作用 [3] [4] [5] 。其中,16S rRNA和COI基因序列均具有较强的种间鉴别能力,已被广泛应用于鱼类的鉴定、分类和系统演化等研究中 [6] [7] [8] 。
本研究通过对比16S rRNA和COI基因部分序列在不同分类阶元中的序列变异,对长江口8种虾虎鱼类早期资源进行种类鉴定和系统进化分析,对比线粒体16S rRNA和COI基因片段在虾虎鱼科鱼类系统分类中的适应性,以期为虾虎鱼科鱼类的物种鉴定和系统分类筛选适当的分子标记。
2. 材料与方法
2.1. 材料
本研究中所用的纹缟虾虎鱼(Tridentiger trigonocephalus)、髭缟虾虎鱼(Tridentiger barbatus)、爪哇拟虾虎鱼(Pseudogobius javanicus)、斑尾刺虾虎鱼(Acanthogobius ommaturus)、睛尾蝌蚪虾虎鱼(Lophiogobius ocellicauda)、拉氏狼牙虾虎鱼(Odontamblyopus lacepedii)、青弹涂鱼(Scartelaos histiophorus)、子陵吻虾虎鱼(Rhinogobius giurinus)等虾虎鱼科鱼类及中国花鲈(Lateolabrax maculatus)和棘头梅童鱼(Collichthys lucidus)均采自长江口水域,并参照《中国动物志》根据可量和可数性状指标对所采集样本进行种类鉴定 [1] 。文中虾虎鱼科共计7属8种24个个体的16S rRNA和COI基因进行同源性分析,并在同一水域采集中国花鲈和棘头梅童鱼,对其序列进行扩增,并通过NCBI (National Center for Biotechnology Information)对序列同源性进行比对,选取2种5个同源序列作为外群,所用样品的分类地位、样本量及编号等相关信息见表1。
2.2. 基因组DNA提取、PCR扩增及序列测定
利用天根海洋动物基因组织提取试剂盒(天根生化科技有限公司,北京)提取基因组DNA。采用16S rRNA通用引物16S AR/16S BR(5'-CGCCTGTTTAACAAAAACAT-3'/5'-CCGGTCTGAACTCAGATCATGT-3')扩增16S rRNA片段 [9] ;采用COI通用引物F/R 5'-TCAACCAACCACAAAGACATTGGCAC-3'/5'-TAGACTTCTGGG TGGCCAAAGAATCA-3')扩增COI基因片段 [10] 。PCR反应总体积为50 μL,其中Premix Taq 25 μL (1.25 U),分别加入引物16S AR/16S BR、F/R各1 μL (20 μmol·L−1),根据纯度和质量加入适量DNA模板,补加双蒸水至50 μL。PCR反应条件为:94℃ 预变性5 min;然后35个循环,包括94℃变性45 s,依据不同引物50.5℃~55℃退火45 s (16S rRNA 52℃,COI 55℃),根据扩增片段长度72℃ 延伸50 s~1 min,最后72℃延伸10 min;PCR产物4℃保存备用。
将扩增产物用1.0%琼脂糖凝胶电泳检测。PCR产物由生工生物工程(上海)股份有限公司进行正反向双链测序。
2.3. 数据分析
将所测序列用NCBI的Blast工具与已知种类鱼类的线粒体基因序列进行比对,确认所得序列为目的片段。应用DNAstar 5.0软件进行拼接,并辅以手工校对。用MEGA 5.0软件对序列进行多重对比,分析统计序列的碱基组成、碱基变异频率、简约信息位点数、单独位点数、转换与颠换位点的比例,进行系

Table 1. Relevant information of samples used in this study
表1. 文中所用样品的相关信息
统分类分析。采用Kinura双参数替代模型(Kimura’s 2-Parameter, K2P)计算种内和种间遗传距离,并用SPSS 19.0软件绘制箱形图,分析16S rRNA和COI基因片段在不同分类阶元中K2P遗传距离的分布情况。建立基于K2P模型的UPGMA系统发生树,采用自展法检验(Bootstrap) 1000次重复检验分子系统树各分支的置信度 [11] 。利用DNAsp 5.1计算其单倍型数目。
3. 结果与分析
3.1. 16S rRNA和COI基因片段的碱基组成
16S rRNA和COI基因片段的碱基组成如表2所示,虾虎鱼科不同种鱼类16S rRNA序列的碱基组成总体趋势基本一致:平均碱基组成分别为T 22.0%,C 24.6%,A 30.2%,G 23.2%,其中碱基A含量最高,其它3个碱基含量相近,平均A + T含量(52.2%)明显高于G + C含量(47.8%)。COI序列的碱基组成总体趋势也基本一致:平均碱基组成分别为T 30.2%,C 27.4%,A 23.9%,G 18.5%,其中碱基T含量最高,其次为碱基C、A、G依次降低,平均A + T含量(54.2%)明显高于G+C含量(45.8%)。
3.2. 16S rRNA和COI基因各密码子的碱基偏向性
密码子3个位点中的碱基组成有显著差异(见表3):虾虎鱼科不同种鱼类16S rRNA序列各密码子的碱基组成基本一致,3个密码子位点的各碱基含量差异较小,在3个密码子间T的变幅为21%~22%,C的变幅为23.4%~26.6%,A的变幅为30.0%~30.4%,G的变幅为20.6%~25.1%。COI序列各密码子的碱基组成差异较大,其中,1st密码子中T含量最多,高达42%,2st密码子中G的含量较为特殊,仅为10.8%,出现了反G偏倚现象,而3st密码子恰好与1 st密码子中碱基含量相反,T的含量最低(18%),而G的含量最高(29.7%),COI序列密码子的碱基使用频率存在明显的偏向性。
3.3. 16S rRNA和COI基因片段的碱基变异
测序获得虾虎鱼科7属8种24个个体的16S rRNA基因片段,比对得到16S rRNA基因长度为535 bp

Table 2. The bases contents of 16S rRNA and COI gene of 8 Gobiidae species
表2. 虾虎鱼科8种鱼类的16S rRNA和COI基因序列的碱基组成

Table 3. Nucleotide frequencies of 16S rRNA and COI partial sequence of 8 Gobiidae species (%)
表3. 8种虾虎鱼科鱼类的16S rRNA和COI基因部分序列中各碱基分布频率(%)
序列,共编码178个氨基酸。24条16S rRNA序列共有12个单倍型,共检测到保守位点374个,变异位点161个(约占30.1%),包括简约信息位点158个,单变位点3个,插入/缺失位点15个;共有26个转换位点,24个颠换位点,转换与颠换比值(Si/Sv)为1.51。测序获得虾虎鱼科7属8种24个个体的COI基因片段,对比得到COI基因长度为641 bp的序列,共编码213个氨基酸。24条COI序列共有12个单倍型,共检测到保守位点420个,变异位点221个(约占34.5%),全部为简约信息位点,没有单变位点,未发现插入/缺失位点;共有58个转换位点,45个颠换位点,转换与颠换比值(Si/Sv)为1.29。8种虾虎鱼的2种基因片段都显示第2密码子位点碱基替换最多,尤其COI基因的第2密码子位点的转换值和颠换值明显高于其它2个密码子位点(表4)。
3.4. 16S rRNA和COI基因的突变饱和分析
分别将16S rRNA和COI基因序列中成对比较推断的碱基替代率和Kumura双参数距离进行比较来
验证虾虎鱼科鱼类各自的基因中是否存在突变饱和现象。线粒体16S rRNA和COI基因序列全部位点的转换率和颠换率与相应的序列差异的散点图如图1所示。图中显示这两个基因序列中的碱基替代率与遗传距离之间均呈现明显的线性关系,因此,它们在虾虎科鱼类中不存在突变饱和现象。
3.5. 遗传距离分析
研究显示,不同物种的16S rRNA和COI序列均各不相同,不同种之间未有序列共享现象。8种虾虎鱼科鱼类种间和种内遗传距离如表5所示,16S rRNA的种内平均K2P遗传距离较小,仅为0.002,而平均种间遗传距离为0.169,约为种内遗传距离的84倍,其中纹缟虾虎鱼和髭缟虾虎鱼的种间遗传距离最小为0.100,拉氏狼牙虾虎鱼和斑尾刺虾虎鱼的遗传距离最大为0.237。相比较而言,8种虾虎鱼科鱼

Table 4. Sequence variations of 16S rRNA and COI gene of 8 Gobiidae species
表4. 8种虾虎鱼科鱼类的16S rRNA和COI基因序列各密码子位点碱基变异

Table 5. Genetic distance pairwise and within-species of 16S rRNA (DOWN) and COI (UP) gene in 8 Gobiidae species
表5. 虾虎鱼科8种鱼类16S rRNA (下)和COI (上)基因的种间和种内遗传距离

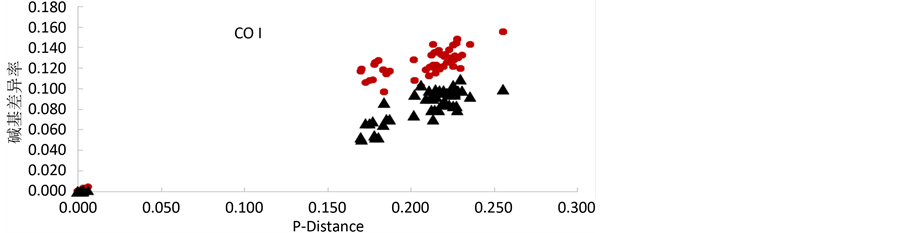

Figure 1. Plot for transition and transversion of the 16S rRNA and COI gene versus P-Distance
图1. 线粒体16S rRNA和COI基因的转换、颠换和P-Distance之间的散点图
类COI序列的种内平均遗传距离仅为0.001,种间遗传距离为0.215,为种内遗传距离的215倍,明显大于16S rRNA序列的种间差异,而COI序列的种间遗传距离最小值是青弹涂鱼与髭缟虾虎鱼之间的遗传距离为0.169,距离最远的是爪哇拟虾虎鱼和纹缟虾虎鱼间的遗传距离为0.263。16S rRNA 和COI基因片段在不同分类阶元中K2P遗传距离的分布情况见图2。除种内16S rRNA的K2P遗传距离略高于COI基因外,同属种间、同科属间及同目科间的16S rRNA基因的K2P遗传距离均明显低于COI基因。
3.6. 虾虎鱼科鱼类的分子系统分类分析
对虾虎鱼科7属8种24个个体与中国花鲈和棘头梅童鱼2个外群的16S rRNA和COI基因序列构建分子进化图谱,结果如图3所示。从图中可明显看出两种基因序列的虾虎鱼科鱼类均形成单系群。在16S rRNA基因序列的亲缘关系图谱中,出现在虾虎鱼科分支基层的为拉氏狼牙虾虎鱼,其次为斑尾刺虾虎鱼,髭缟虾虎鱼和纹缟虾虎鱼构成姐妹分支(置信度为90%),说明二者亲缘关系较近(图3(a));而在COI基因序列的亲缘关系图谱中,出现在虾虎鱼科分支基层的是爪哇拟虾虎鱼,其次为斑尾刺虾虎鱼和睛尾蝌蚪虾虎鱼,并且斑尾刺虾虎鱼和睛尾蝌蚪虾虎鱼形成姊妹分支,其置信度为83%,表明二者具有较近的亲缘关系(图3(b))。
4. 讨论
已有研究表明线粒体不同基因的进化速率存在差异 [11] [12] [13] 。在线粒体基因组中16s rRNA基因较为保守,进化速率较慢,而COI基因的变异性则相对较大 [14] [15] [16] [17] 。本研究中在虾虎鱼科8种24个个体的16S rRNA基因序列中共检测到161个碱基变异,约占30.1%;而在COI序列中发现变异位点221个,占34.5%,可见COI基因进化速率相对16S rRNA基因较快,进一步证实16S rRNA基因相对保守这一结论。同时,本研究中16S rRNA和COI基因序列全部转换与颠换个数的比值分别为1.51和1.29,均小于2。但通过对其进行碱基突变饱和性分析,虾虎鱼科鱼类基因序列的碱基替代速率同遗传距

Figure 2. Box chart of distribution of K2P distance for the two mtDNA genes fragments within different taxonomic levels
图2. 2种基因在不同分类阶元的K2P遗传距离分布箱型图
 (a)
(a) (b)
(b)
Figure 3. UPMGA phylogenetic tree of Gobiidae inferred from 16S rRNA and COI gene sequences
图3. 基于16S rRNA (a)和COI (b)部分基因序列构建虾虎鱼科鱼类的UPGMA树
离进行比较的结果显示这两种基因中无论转换还是颠换都没有达到饱和,因此它们能够用于虾虎鱼科鱼类的分子系统分类分析,该结果与孙希褔等 [18] 对10种虾虎鱼类的研究结果一致。
Asakawa等 [19] 提出后生动物线粒体基因的碱基构成具有明显的偏倚性,通常表现为A + T含量显著高于G + C含量。本研究8种虾虎鱼科鱼类的16S rRNA和COI序列的A+T含量均明显高于G + C含量,进一步验证了上述偏倚性的特征 [15] [16] [17] [18] [19] 。Saccone等 [20] 分析后生动物的线粒体基因组全序列得出,硬骨鱼类的平均G + C含量(43.2%)显著高于软骨鱼类(38.4%)。本研究中虾虎鱼科鱼类的16S rRNA和COI基因序列平均G + C含量分别为47.8%和45.8%,均显著高于软骨鱼类的平均G + C含量,符合硬骨鱼类的序列组成特征。此外,COI基因序列密码子3个位点中的碱基组成有显著差异,表明COI序列密码子间的碱基使用频率存在明显的偏向性。
从遗传距离角度分析,种内和种间遗传距离的大小是进行种类鉴定的重要依据。Hebert等 [21] 认为COI序列在物种间的遗传差异通常大于0.02,并提出当种间遗传距离与种内遗传距离的差异达到10倍以上时,COI基因才可对物种进行有效的鉴定,这一观点已在许多物种的研究中得到了验证 [22] [23] [24] 。本研究通过对比分析16S rRNA和COI基因序列,得出种内平均遗传距离分别为0.002和0.001,种间平均遗传距离分别为0.169和0.215,种间遗传距离远大于种内遗传距离,因此,16S rRNA和COI基因序列均能有效地对长江口虾虎鱼科鱼类进行物种鉴定。但从表5可以看出,16S rRNA序列间的种间平均遗传距离明显小于COI序列,由此可见,COI基因在种间的趋异性要明显高于16S rRNA基因,进一步说明在本研究的虾虎鱼科鱼类中,COI基因的进化速率相对较快,16S rRNA基因则相对保守些。
本研究分子系统图谱的拓扑结构显示,两种基因序列的虾虎鱼科鱼类均为明显的单系群,其中虾虎鱼类的分支中包括了以传统分类标准划分的弹涂鱼科的青弹涂鱼以及鳗虾虎鱼科的拉氏狼牙虾虎鱼 [25] 。从亲缘关系来看,上述两种鱼类与其它虾虎鱼类的亲缘关系密切,均可归为虾虎鱼科。该划分与伍汉霖等 [1] 主编的《中国动物志》中对以上两种鱼类的分类结果一致。在16S rRNA基因的亲缘关系图谱中显示,髭缟虾虎鱼和纹缟虾虎鱼形成姊妹分支,其遗传距离只有0.100,说明两者亲缘关系较近,同属于缟虾虎鱼属 [1] ;而在COI基因的亲缘关系图谱中显示,亲缘关系较近的为睛尾蝌蚪虾虎鱼和斑尾刺虾虎鱼,二者间的遗传距离也最小,仅为0.189。该结果与于亚男等 [2] 的研究结果相符,但在基于COI亲缘关系图谱中除了髭缟虾虎鱼与纹缟虾虎鱼聚在一起外,该分支中还包括青弹涂鱼、拉氏狼牙虾虎鱼和子陵吻虾虎鱼,表明这5种虾虎鱼在COI基因中近缘关系相对较近,不能把不同属的虾虎鱼进行有效区分。可见,16S rRNA基因较COI基因在虾虎鱼属水平上有更好的种类分辨能力。
据相关文献报道,已有许多学者利用16S rRNA和COI基因部分序列对多种物种进行了种间系统进化关系分析,将两种基因在同一物种中进行适用性的比较等相关研究 [17] [26] [27] [28] [29] 。本研究通过对线粒体16S rRNA和COI基因片段的遗传特性及其在虾虎鱼科鱼类种类鉴定和系统分类中的适用性进行对比,结果表明,在长江口虾虎鱼科鱼类中,16S rRNA基因序列的碱基变异率明显低于COI基因,进一步证实了COI基因的进化速率相对较快,16S rRNA基因则相对保守这一结论。有研究表明16S rRNA基因在一些近缘种的区分上存在困难,不适用于种水平的物种鉴定,然而在另外一些动物类群中,16S rRNA基因却被证明有相对较高的变异水平 [14] [30] 。本研究表明,16S rRNA基因和COI基因序列均能有效地对长江口虾虎鱼科鱼类进行物种鉴定,但相对于COI基因来说,16S rRNA基因在虾虎鱼科属的水平上有较好的物种鉴别能力,因此,在应用这两种基因片段进行系统分类研究时,应根据研究的不同系统水平选择适当的分子标记。
基金项目
东海水产种质资源标准化整理、整合与共享。