1. 引言
每个细胞信号的传递都是借助细胞膜的不同种类的受体,将细胞外的信号传递到细胞内。G蛋白偶联受体(G Protein-Coupled Receptors, GPCRs)就是一个因能结合和调节G蛋白活性而得名的超级膜蛋白家族。作用于GPCRs的信号物质,通过影响细胞的GPCRs而对G蛋白质起作用。因此GPCRs被认为是相似的分子机制而起作用。GPCRs在信号传导中的重要作用,不仅有助于了解细胞信号的传导机制、阐明疾病的致病机理,而且对药物的研究提供新的思路,GPCRs的功能失调会引发许多疾病,如疼痛、色盲症、哮喘等。通过调节有关GPCRs介导的信号传导,可以治疗高血压、紧张和消化道溃疡等病症。大部分药物可通过靶向作用于GPCRs而达到治疗的效果,所以GPCRs在制药领域成为重要的药物作用靶标。根据GPCRs的序列差异,准确地聚类GPCRs序列有着很重要的理论意义和应用价值 [1] [2] 。
蛋白质序列相似性分析是蛋白质序列聚类的关键所在。经典的相似性算法有Needleman-Wunsch算法、Smith-Waterman算法、接触度量矩阵法、矩阵法、T-coffee算法、SIM算法、基于氨基酸物化性的拓扑指数方法和基于LZ复杂度等方法 [3] [4] [5] [6] 。尽管这些算法都有它们各自的优点,但是计算量都比较大。在序列比对算法中,仅用相同或不同来说明两个残基之间的关系,无法描述残基替换对结构和功能的影响程度;矩阵法是构造一个适当的数值矩阵来描述蛋白质序列,然后选取矩阵的不变量把蛋白质序列之间的比较转化成矩阵不变量之间的比较,并且把初始的蛋白质字符串序列转换为特征向量,这些特征向量的维数可以按照自己的愿望进行选择。通过对这些不变量的比较来确定蛋白质序列的相似与不相似 [7] - [12] 。虽然这个方法简洁快速,但是在蛋白质序列转化成特征向量的过程中,总会丢失了一些生物信息;同样在LZ复杂度法中也只考虑了结构信息而忽略了氨基酸的物化性质对蛋白质的结构和功能的影响 [13] ,为此促使人们试图寻找其它更有效的方法来比较蛋白质序列的相似性。
本文中,首先在氨基酸的物化性质表征蛋白质序列的基础上,把蛋白质序列转化成11维的特征向量;其次,根据20种氨基酸的极性、非极性、疏水性、亲水性将其分为四类:极性且亲水性
、极性且疏水性
、非极性且亲水性
和非极性且疏水性
,将这四类氨基酸两两连接得到16个特征子列,并计算了16个特征子列在蛋白质序列中出现的频率,利用此频率将蛋白质序列转化成16维特征向量;最后,用因子分析法把蛋白质序列的特征向量进行降维得到因子模型,进而利用因子模型分析40个G蛋白偶联受体序列的相似性,并对其进行聚类分析。
2. 因子模型
是可观测的随机向量,
,
,
是不可观测的随机向量,
,
,又设
与
不相关,且
,
。设
满足:
(2.1)
矩阵方程
称正交因子模型,其中
称
的公共因子,
称
的特殊因子。设
分别是均值为0,方差为1的随机变量,即
;特殊因子
分别是均值为0,方差为
的随机变量,即
;各特殊因子之间及特殊因子与公共因子之间都是相互独立的,即
及
。
是第
个变量在第
个公共因子上的负荷,
是待估系数矩阵(因子载荷矩阵) [14] 。
因子分析的目标是找出公共因素及特有的因素,即公共因子与特殊因子。在公因子分析中,特殊因子起到残差的作用,但被定义为彼此不相关且和公因子也不相关。而且每个公因子假定至少对两个变量有贡献,否则它将是一个特殊因子。在开始提取公因子时,为了简便,还假定公因子彼此不相关且具有单位方差。在这种情况下,向量
的协方差矩阵
可以表示为:
(2.2)
这里
,
表示对角矩阵。
如果已知
协方差矩阵
和
,可以很容易地求出
。根据式(2.2)有:
记
,则
是非负定矩阵。若记矩阵
的
个特征值
且
,及
分别表示m个非零特征值所对应的标准化的特征向量,则
的谱分解式为:
(2.3)
只要:
(2.4)
就可以求出因子载荷矩阵
。从而求得
,这时
和
为因子模型的一个解,这个解就称为主因子解。
因子模型被估计后,还必须对得到的公因子
给出一种明确的解释,它用来反映在预测每个可观察变量中这个公因子的重要性,这个公因子的重要程度就是在因子模型矩阵中相应于这个因子的系数,显然这个因子的系数绝对值越大越重要,而接近0则表示对可观察变量没有什么影响。因子解释是一种主观的方法,有时侯,通过旋转公因子可以减少这种主观性,也就是要使用非奇异的线性变换。
设
维可观察变量
满足因子模型
,设
是任一正交阵,则因子模型可改写为:
(2.5)
其中,
,
。
3. 蛋白质序列的特征向量表示
3.1. 蛋白质序列的11维、2维特征向量表示
根据大量的实验结果,我们提取了对G偶联受体序列相似性分析有影响的氨基酸的11种物化属性 [15] ,它们是:
(
螺旋的
端动力),
(
螺旋接触面积),
(可压缩性),
(
折叠趋势),
(在溶剂中的收缩率),
(溶剂可及表面积),
(氨基酸的等电点),
(吉布斯自由能变性蛋白水化的变化),
(平均中程接触),
(折射率),
(长距离的非键能),并对其进行了标准化和平均化 [15] ,见表1。
根据20种氨基酸的标准化和平均化后的物化属性,对40个G蛋白受体序(https://www.ncbi.nlm.nih. gov/protein/1LMB_4上下载)中所含的20个氨基酸进行统计并计算它们的算数平均数,由于数据篇幅比较大,表2只给出了部分G蛋白受体序列的算术平均数。

Table 1. 20 properties of amino acids
表1. 20种氨基酸的属性取值表

Table 2. The arithmetic mean of 40 proteins
表2. 40种蛋白质的算数平均值
根据得到蛋白质序列对应的11维特征向量,并计算11维特征向量的相关矩阵及其特征值,根据特征值累计贡献率提取相应的主因子,以主因子的方差贡献率作为权重得到因子模型如下:
,
,
,
,
,
,
,
,
,
,
其中
表示
,
表示
,
表示
,
表示
,
表示
,
表示
,
表示
,
表示
,
表示
,
表示
,
表示
,
是
的标准化,利用因子模型可得蛋白质序列对应的2维特征向量
。例如,蛋白质序列Q8MXU2对应的二维特征向量
。
3.2. 蛋白质序列的16维、6维特征向量表示
根据20种氨基酸的极性、非极性、疏水性、亲水性将其分为四类 [16] :极性且亲水性
、极性且疏水性
、非极性且亲水性
和非极性且疏水性
,将这四类氨基酸两两连接得到16个特征子列:
,
,
,
,
,
,
,
,
,
,
,
,
,
,
,
,并计算这些特征子列在蛋白质序列中出现的频率得到蛋白质序列的16维特征向量表示。在此基础上,根据得到蛋白质序列对应的16维特征向量,并计算16维特征向量的相关矩阵及其特征值,根据特征值累计贡献率提取相应的主因子,以主因子的方差贡献率作为权重得到因子模型。利用因子模型可得蛋白质序列对应的6维特征向量
。例如,蛋白质序列Q8MXU2对应的二维特征向量
。
4. 蛋白质序列相似性及其聚类分析
从https://www.ncbi.nlm.nih.gov/protein/1LMB_4上下载了40个G蛋白受体序列 [17] 。下面采用向量端点之间的平方和距离来构造相似性矩阵。由于数据过多,40个G蛋白偶联受体序列对应的相似性矩阵不列在本文里。把相似性矩阵输入到SAS软件得到了40个G蛋白受体序列的聚类图,如图1~图4所示。
观察聚类图1~图4易看出,40个G蛋白偶联受体序列中:P97772和Q9UGT0,O00222和Q3MIV9,Q93564和Q622H2,Q6ZMQ2和Q14833,P31421和Q14416,Q5RAL3、Q9QYS2和P31422,Q863I4、O15303和P35349,Q93564和Q622H2最相似。根据文献 [13] ,如Q68EF4和Q14833,P47743和P70579,Q9V4U4和Q70GQ8,Q5TZ45和P31424是比较相似的。而通过观察图我们发现,只有16维向量和6维向量所得到的聚类图满足,11维向量和2维向量的到的聚类图只满足一部分。由此可见,根据氨基酸的极性和亲水性得到的蛋白质相似性比较更加合理。而图2和图4是利用因子分析法,把40个G蛋白偶联受体序列对应的11维和16维特征向量降维到2维和6维得到的聚类图。在图2中Q75QW7,Q90ZF3,Q4RJZ9,Q68EF4,Q9V4U4,Q8NHA9,Q4R3P0的位置和图1的有差别。但是通过参考文献 [13] ,我们知道Q75QW7应该和Q4REZ5相近,Q90ZF3、Q4RJZ9和Q4REI8相近,Q14833和Q68EF4相近,Q8NHA9和O15303相近。在图4中Q8MXU2,Q75QW7,Q5RDQ8,Q62916的位置和图3的有差别。而在参考文献 [13] 中,Q75QW7和P91685相近,Q5RDQ8
和Q62916、Q14833和Q68EF4相近。由此可见,基于2维和6维特征向量聚类比11维和16维特征向量聚类效果较好。再把图2与图4与文献 [13] 比较可知,图4与文献 [13] 的结果更一致,这说明,我们选取的20种氨基酸的极性和亲水性对G蛋白偶联受体序列相似性比较中有一定的影响,总之,用低维特征向量聚类过程简单,时间复杂度低等优势,而且实例验证,基于2维特征向量聚类效果也较好。

Figure 1. Cluster analysis of 40 G protein-coupled receptor sequences based on 11-dimensional factor vector
图1. 基于11维特征向量的40个G蛋白偶联受体序列聚类图

Figure 2. Cluster analysis of 40 G protein-coupled receptor sequences based on 2-dimensional factor vector
图2. 基于2维特征向量的40个G蛋白偶联受体序列聚类图
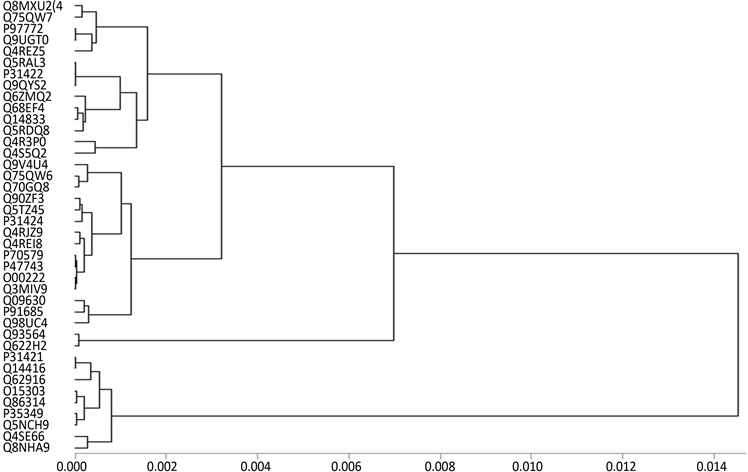
Figure 3. Cluster analysis of 40 G protein-coupled receptor sequences based on 16-dimensional factor vector
图3. 基于16维特征向量的40个G蛋白偶联受体序列聚类图
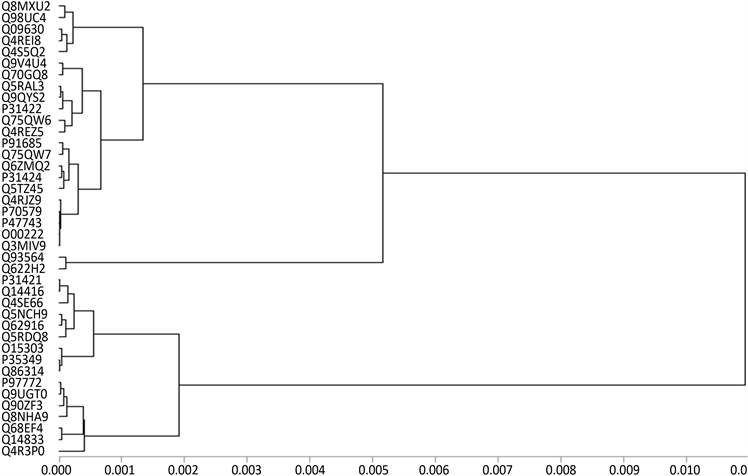
Figure 4. Cluster analysis of 40 G protein-coupled receptor sequences based on 6-dimensional factor vector
图4. 基于6维特征向量的40个G蛋白偶联受体序列聚类图
5. 总结
因子分析是主成分分析的推广,它是一种降维方法,其目的是用有限个不可观测的隐变量来解释原始变量之间的相关性。在因子模型中,由于因子载荷矩阵不是唯一的,我们利用这一特点可以通过因子的旋转,使得旋转后的因子有更鲜明的实际意义。本文利用因子分析法,对40个G蛋白偶联受体序列进行相似性分析,并将其聚类,得到的结果验证了本方法的可行性。而且,此方法对生物序列进化的研究具有操作简单、时间复杂度低、对序列的长度没有限制等优点。对于本文中我们选取的氨基酸的11种物化性质在不同功能的蛋白质中有没有规律以及分析这些物化性质与蛋白质功能和结构之间的影响都是有待于进一步研究的课题。
基金项目
感谢基金项目:辽宁省教育厅科学研究一般项目(No.L 2015093)对本论文的支持。同时,也要衷心地感谢本文中引用文章的作者。